

作者: 天诺基业时间:2021-04-06
适合稳定同位素混合模型(SIMMs),可以长期替代以前广泛使用的SIAR包,包含了不确定性、浓度依赖,及大量来源分析。SIMMs是通过观察从生物体组织样本中获取的稳定同位素值来推断生物体消费各种食物来源的膳食比例。然而,SIMMs也可用于其他情况,例如在沉积物混合或脂肪酸组成中。主要函数是simmr_load和simmr_mcmc。这两个小节包含了一个快速开始和所有功能的完整列表。
一种常见的应用是利用消费者及其食物的稳定同位素组成来推断动物的同化食物的组成。
依赖
R(版本>=3.5.0),R2jags,ggplot2
simmr包使用JAGS(另一种吉布斯采样器)程序来运行稳定同位素混合模型。在安装simmr之前,请访问JAGS网站并下载并安装适合您的操作系统的JAGS。
编码
UTF-8
内置数据
TRUE

描述
这个函数接受simmr_output类的对象,并组合两个食物源。它适用于单个和多个组数据。
用法
1combine_sources(
2 simmr_out,
3 to_combine = simmr_out$input$source_names[1:2],
4 new_source_name = "combined_source"
5 )
输入参数

详细
通常两个源要么 (1) 在同位素空间图上位于相似位置,要么 (2) 在系统发育方面非常相似。在 (1) 中,来源之间的后验相关性通常很高(负)。将它们结合起来可以减少这种相关性并提高估计的精度。在 (2) 中,我们可能希望确定这两种食物合并后的共同食用量。因此,该函数在运行simmr_mcmc(称为后验组合)之后组合了两个源。然后可以使用plot.simmr_input或plot.simmr_output调用此新对象,生成组合后输出的同位素空间图概要。
输出值
为一个新的simmr_output对象
示例
1library(rjags)
2library(R2jags)
3library(ggplot2)
4library(simmr)
simmr包内自带的数据集1print(data(package='simmr'))
Data sets in package simmr:

1help(geese_data_day1)
geese_data_day1是一个真实的布兰特鹅数据集,包括对2种同位素(d13C、d15N)、4种来源(Zostera、Grass、U.lactuca、Enteromorpha)、校正/营养富集因子(TEFs或TDFs)和浓度依赖均值的9次观测。
with函数,通过simmr_load方法加载simmr包自带的geese_data_day1数据集中的数据到结构体simmr_1 1data(geese_data)
2# Load into simmr
3simmr_1 <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
1# Plot
2plot(simmr_1)

simmr_11#Print
2simmr_1
This is a valid simmr input object with 9 observations, 2 tracers, and 4 sources.
The source names are: Zostera, Grass, U.lactuca, Enteromorpha.
The tracer names are: d13C_Pl, d15N_Pl.
simmr_mcmc1# MCMC run
2simmr_1_out <- simmr_mcmc(simmr_1)
module glm loaded
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 18
Unobserved stochastic nodes: 6
Total graph size: 128
Initializing model
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
|**************************************************| 100%
simmr_1_out1# Print it
2print(simmr_1_out)
This is a valid simmr input object with 9 observations, 2 tracers, and 4 sources.
The source names are: Zostera, Grass, U.lactuca, Enteromorpha.
The tracer names are: d13C_Pl, d15N_Pl.
The input data has been run via simmr_mcmc and has produced 3600 iterations over 4 MCMC chains.
1# Summary
2summary(simmr_1_out)
3summary(simmr_1_out, type = "diagnostics")
4summary(simmr_1_out, type = "correlations")
5summary(simmr_1_out, type = "statistics")
6ans <- summary(simmr_1_out, type = c("quantiles", "statistics"))
1#plot
2plot(simmr_1_out)
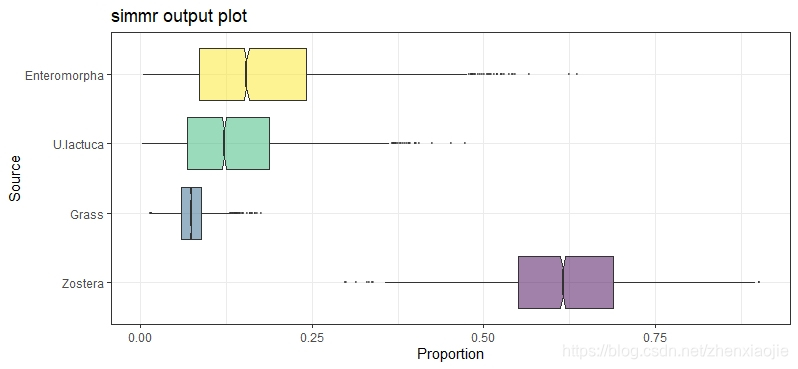
1plot(simmr_1_out, type = "boxplot")
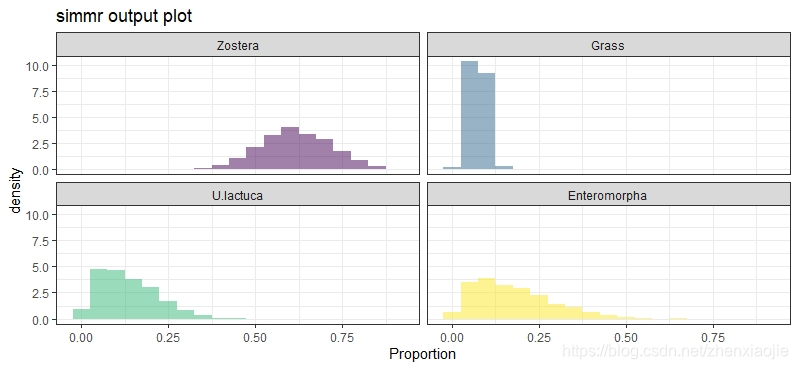
1plot(simmr_1_out, type = "histogram")
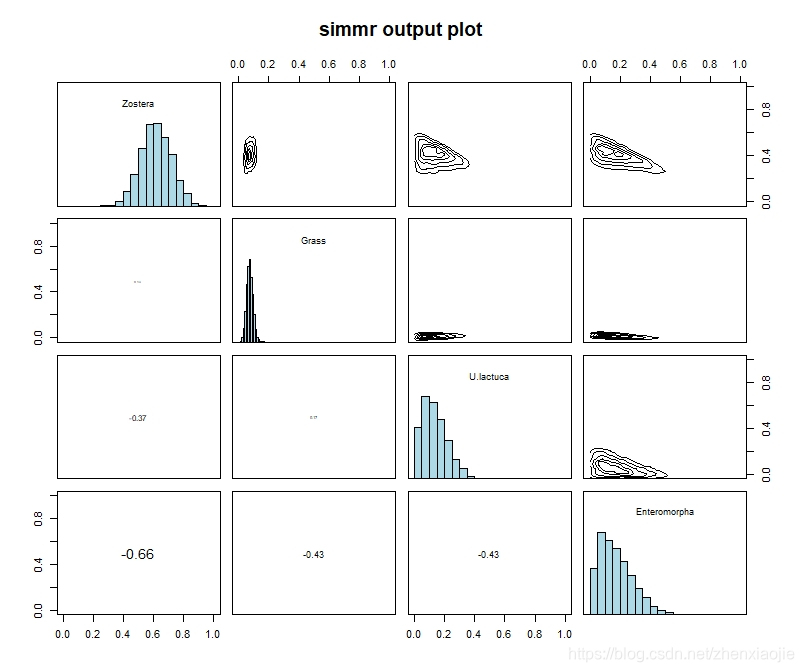
1plot(simmr_1_out, type = "matrix")
1plot(simmr_1_out, type = "matrix")
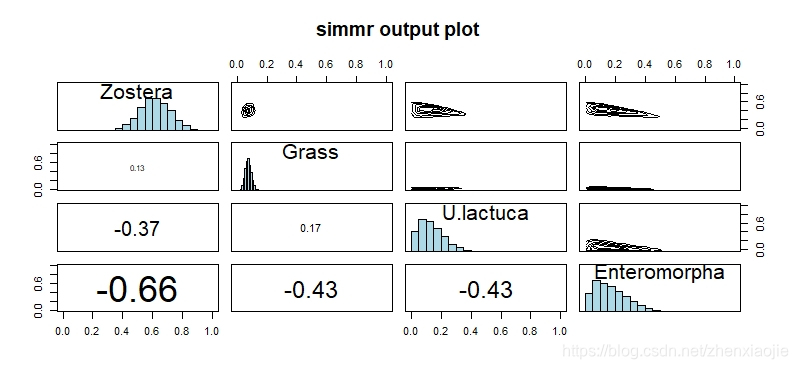
simmr_mcmc的输出带入combine_sources,将在同位素空间分布接近的源合并,生成新的组合源1simmr_out_combine <- combine_sources(simmr_1_out,
2 to_combine = c("U.lactuca", "Enteromorpha"),
3 new_source_name = "U.lac+Ent"
4)
1plot(simmr_out_combine$input)
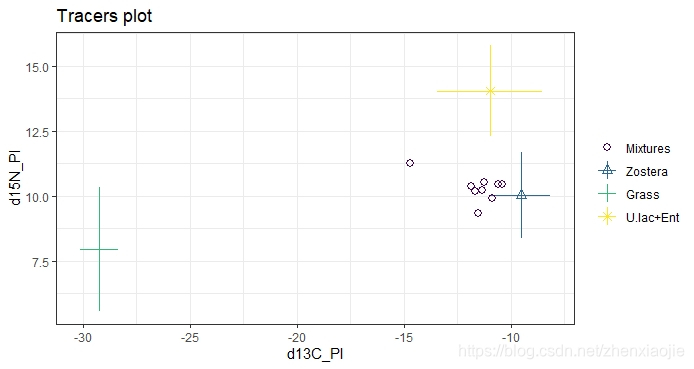
1plot(simmr_out_combine, type = "boxplot", title = "simmr output: combined sources")


描述
compare_groups函数接受simmr_output类的对象,并为给定的源和至少两个组的集合创建概率比较
用法
1compare_groups(
2 simmr_out,
3 source_name = simmr_out$input$source_names[1],
4 groups = 1:2,
5 plot = TRUE
6)
输入参数

详细
当指定了两组时,函数会直接计算其中一组比另一组大的概率。当给出两个以上的组时,该函数为每个组的组合生成一组最有可能的概率排序。该函数在默认情况下生成箱线图,如果需要,还允许存储输出以供进一步分析。
输出值
如果有两组,输出值为一个包含该源在两组间膳食比例差异的向量。如果有多个组,输出值为一个包含以下字段的列表

示例
simmr数据集中的geese_data到simmr_in 1## Not run:
2data(geese_data)
3simmr_in <- with(
4 geese_data,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means,
13 group = groups
14 )
15)
simmr_in1# Print
2simmr_in
This is a valid simmr input object with 251 observations, 2 tracers, and 4 sources. There are 8 groups.
The source names are: Zostera, Grass, U.lactuca, Enteromorpha.
The tracer names are: d13C_Pl, d15N_Pl.
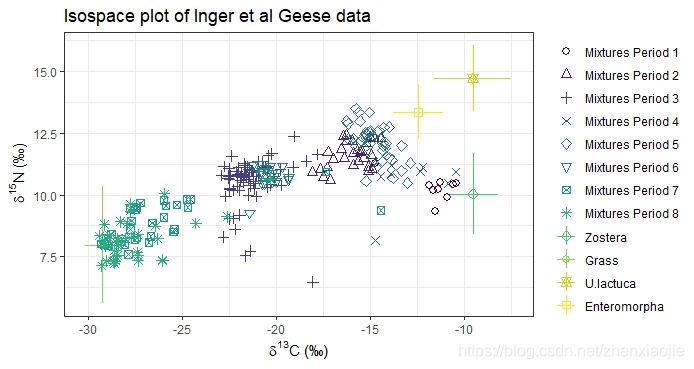
1# Plot
2plot(simmr_in,
3 group = 1:8, xlab = expression(paste(delta^13, "C (\u2030)", sep = "")),
4 ylab = expression(paste(delta^15, "N (\u2030)", sep = "")),
5 title = "Isospace plot of Inger et al Geese data"
6)
simmr_mcmc,并打印模型运行概要1# Run MCMC for each group
2simmr_out <- simmr_mcmc(simmr_in)
3# Print output
4simmr_out
This is a valid simmr input object with 251 observations, 2 tracers, and 4 sources. There are 8 groups.
The source names are: Zostera, Grass, U.lactuca, Enteromorpha.
The tracer names are: d13C_Pl, d15N_Pl.
The input data has been run via simmr_mcmc and has produced 3600 iterations over 4 MCMC chains.
1# Summarise output
2summary(simmr_out, type = "quantiles", group = 1)
3summary(simmr_out, type = "quantiles", group = c(1, 3))
4summary(simmr_out, type = c("quantiles", "statistics"), group = c(1, 3))
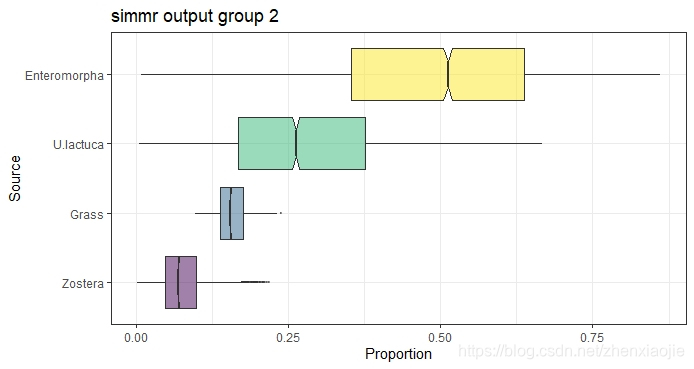
1# Plot - only a single group allowed
2plot(simmr_out, type = "boxplot", group = 2, title = "simmr output group 2")
1plot(simmr_out, type = c("density", "matrix"), grp = 6, title = "simmr output group 6")
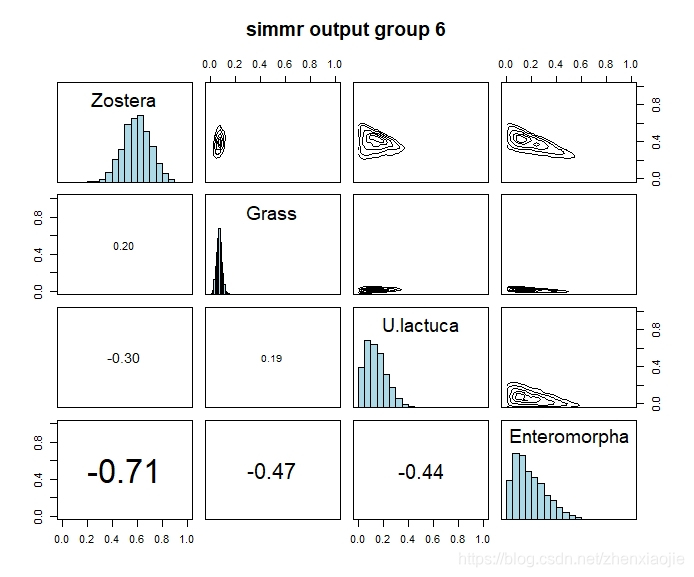
Zostera的膳食比例对比1# Compare groups
2compare_groups(simmr_out, source = "Zostera", groups = 1:2)
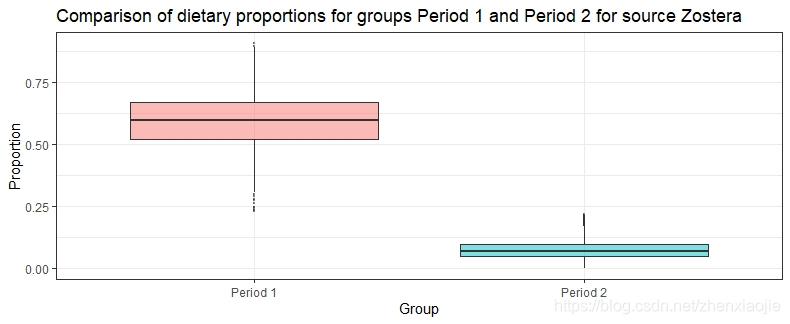
Prob ( proportion of Zostera in group Period 1 > proportion of Zostera in group Period 2 ) = 1
Zostera的膳食比例对比1compare_groups(simmr_out, source = "Zostera", groups = 1:3)
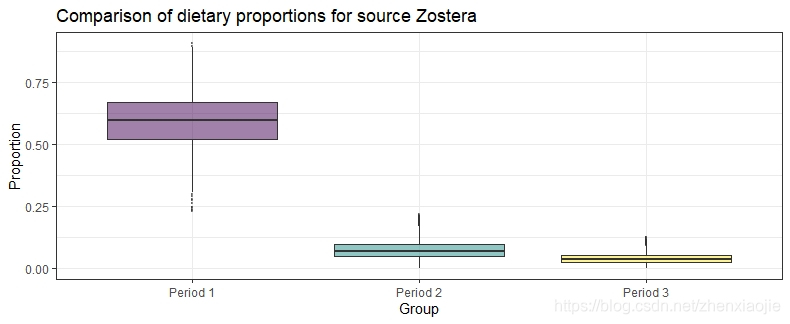
Most popular orderings are as follows:
Probability
Period 1 > Period 2 > Period 3 0.8019
Period 1 > Period 3 > Period 2 0.1981
U.lactuca的膳食比例对比1compare_groups(simmr_out, source = "U.lactuca", groups = c(4:5, 7, 2))

Most popular orderings are as follows:
Probability
Period 2 > Period 5 > Period 4 > Period 7 0.1825
Period 5 > Period 2 > Period 4 > Period 7 0.1394
Period 5 > Period 4 > Period 2 > Period 7 0.1072
Period 2 > Period 4 > Period 5 > Period 7 0.1003
Period 2 > Period 5 > Period 7 > Period 4 0.0928
Period 5 > Period 2 > Period 7 > Period 4 0.0792
Period 4 > Period 5 > Period 2 > Period 7 0.0642
Period 4 > Period 2 > Period 5 > Period 7 0.0481
Period 5 > Period 4 > Period 7 > Period 2 0.0442
Period 4 > Period 5 > Period 7 > Period 2 0.0231
Period 5 > Period 7 > Period 2 > Period 4 0.0228
Period 2 > Period 4 > Period 7 > Period 5 0.0219
Period 5 > Period 7 > Period 4 > Period 2 0.0169
Period 2 > Period 7 > Period 5 > Period 4 0.0153
Period 4 > Period 2 > Period 7 > Period 5 0.0097
Period 2 > Period 7 > Period 4 > Period 5 0.0072
Period 4 > Period 7 > Period 2 > Period 5 0.0061
Period 4 > Period 7 > Period 5 > Period 2 0.0053
Period 7 > Period 5 > Period 4 > Period 2 0.0042
Period 7 > Period 5 > Period 2 > Period 4 0.0033
Period 7 > Period 4 > Period 2 > Period 5 0.0019
Period 7 > Period 4 > Period 5 > Period 2 0.0019
Period 7 > Period 2 > Period 5 > Period 4 0.0014
Period 7 > Period 2 > Period 4 > Period 5 0.0011

描述
这个函数接受simmr_output类的对象,并在提供的源之间创建概率比较。也可以指定组号。
用法
1compare_sources(
2 simmr_out,
3 source_names = simmr_out$input$source_names,
4 group = 1,
5 plot = TRUE
6)
输入参数

详细
当两个来源时,该函数直接计算出一个来源的膳食比例大于另一个来源的概率。当给出两个以上的源时,函数为每个源的组合产生一组最可能的概率排序。该函数在默认情况下生成箱线图,如果需要,还允许存储输出以供进一步分析。
输出值
如果有两个来源,输出值为一个向量,包含两种来源的膳食比例差异。如果有多个源,输出值为一个包含以下字段的列表:

示例
1data(geese_data_day1)
2simmr_1 <- with(
3 geese_data_day1,
4 simmr_load(
5 mixtures = mixtures,
6 source_names = source_names,
7 source_means = source_means,
8 source_sds = source_sds,
9 correction_means = correction_means,
10 correction_sds = correction_sds,
11 concentration_means = concentration_means
12 )
13)
14# Plot
15plot(simmr_1)
16# Print
17simmr_1
18# MCMC run
19simmr_1_out <- simmr_mcmc(simmr_1)
20# Print it
21print(simmr_1_out)
22# Summary
23summary(simmr_1_out)
24summary(simmr_1_out, type = "diagnostics")
25summary(simmr_1_out, type = "correlations")
26summary(simmr_1_out, type = "statistics")
27ans <- summary(simmr_1_out, type = c("quantiles", "statistics"))
28# Plot
29plot(simmr_1_out, type = "boxplot")
30plot(simmr_1_out, type = "histogram")
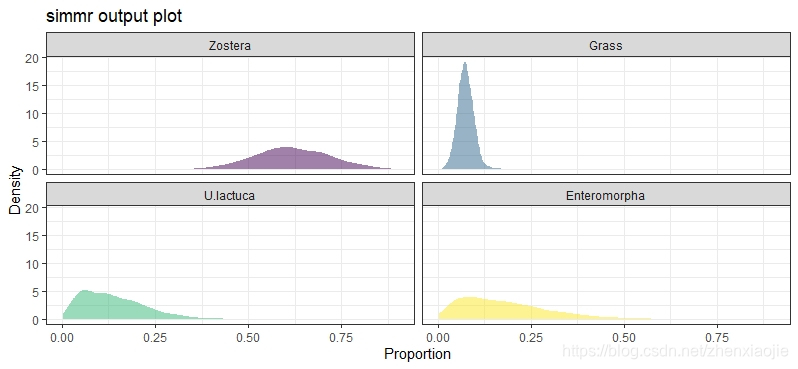
31plot(simmr_1_out, type = "density")
32plot(simmr_1_out, type = "matrix")
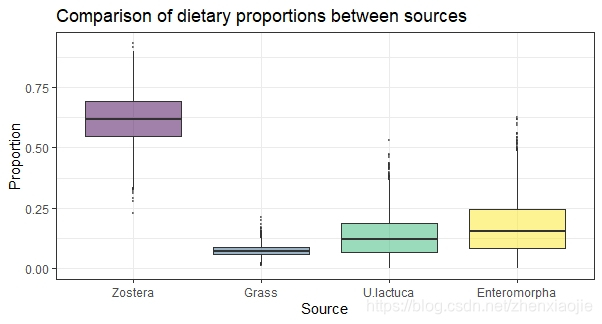
1# Compare two sources
2compare_sources(simmr_1_out, source_names = c("Zostera", "Grass"))

1# Compare multiple sources
2compare_sources(simmr_1_out)

描述
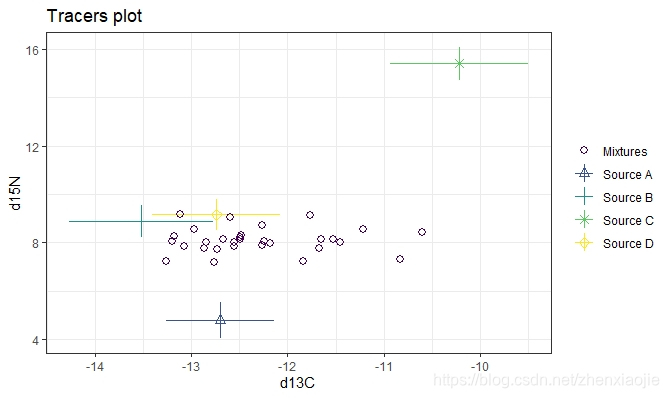
此函数创建同位素空间(又名示踪空间或delta空间)图。它们在确定数据是否适合在SIMMR中运行时至关重要。
用法
1## S3 method for class 'simmr_input'
2plot(
3 x,
4 tracers = c(1, 2),
5 title = "Tracers plot",
6 xlab = colnames(x$mixtures)[tracers[1]],
7 ylab = colnames(x$mixtures)[tracers[2]],
8 sigmas = 1,
9 group = 1:x$n_groups,
10 mix_name = "Mixtures",
11 ggargs = NULL,
12 colour = TRUE,
13 ...
14)
输入参数

详细
理想的情况是将绝大多数的混合物观测值置于由食物源确定的凸壳内。当有两个以上的示踪剂时(如下面的一个例子),建议绘制出所有不同的食物来源对。有关更丰富的绘图细节,请通过vignette('simmr')参阅帮助文档。
示例
1# A simple example with 10 observations, 4 food sources and 2 tracers
2data(geese_data_day1)
3simmr_1 <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15# Plot
16plot(simmr_1)
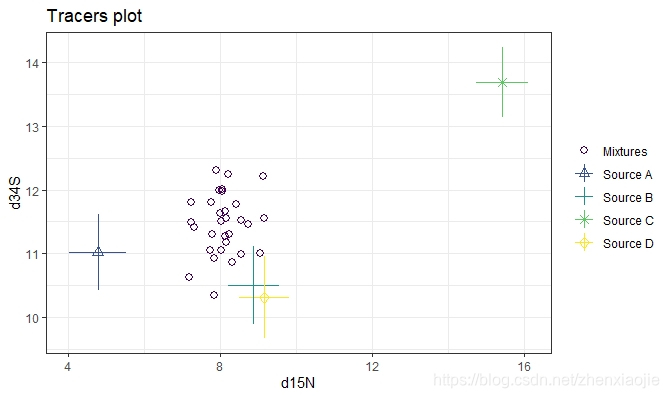
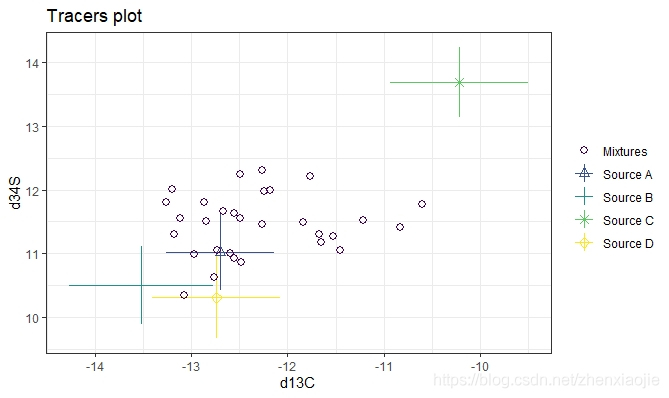
1### A more complicated example with 30 obs, 3 tracers and 4 sources
2data(simmr_data_2)
3simmr_3 <- with(
4 simmr_data_2,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15# Plot 3 times - first default d13C vs d15N
16plot(simmr_3)
17# Now plot d15N vs d34S
18plot(simmr_3, tracers = c(2, 3))
19# and finally d13C vs d34S
20plot(simmr_3, tracers = c(1, 3))
21# See vignette('simmr') for fancier x-axis labels
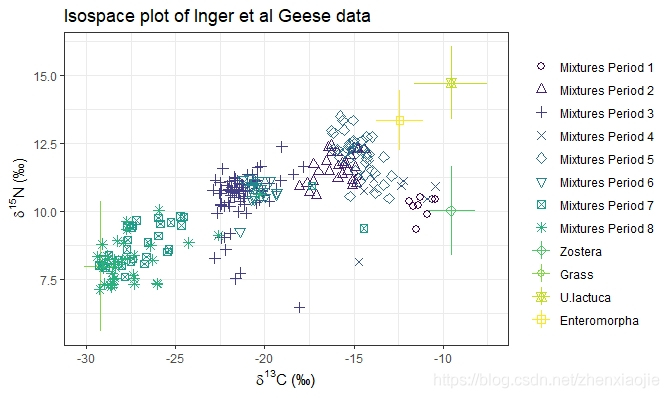
1# An example with multiple groups - the Geese data from Inger et al 2006
2data(geese_data)
3simmr_4 <- with(
4 geese_data,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means,
13 group = groups
14 )
15)
16# Print
17simmr_4
18# Plot
19plot(simmr_4,
20 xlab = expression(paste(delta^13, "C (\u2030)", sep = "")),
21 ylab = expression(paste(delta^15, "N (\u2030)", sep = "")),
22 title = "Isospace plot of Inger et al Geese data"
23) #'

描述
这个函数允许从simmr_mcmc创建4种不同类型的simmr输出图。类型有:直方图、核密度图、矩阵图(最有用)和箱线图。有一些较小的自定义选项。
用法
1## S3 method for class 'simmr_output'
2plot(
3 x,
4 type = c("isospace", "histogram", "density", "matrix", "boxplot"),
5 group = 1,
6 binwidth = 0.05,
7 alpha = 0.5,
8 title = if (length(group) == 1) { "simmr output plot" } else {
9 paste("simmr output plot: group", group) },
10 ggargs = NULL,
11 ...
12)
输入参数

详细
矩阵图应该是任何SIMM分析的必要部分,因为它允许用户判断模型可以识别哪些源。有关更丰富的绘图细节,请通过vignette('simmr')参阅帮助文档。
示例
combine_sources函数介绍中所示,不再赘述。 1# A simple example with 10 observations, 2 tracers and 4 sources
2# The data
3data(geese_data)
4# Load into simmr
5simmr_1 <- with(
6 geese_data_day1,
7 simmr_load(
8 mixtures = mixtures,
9 source_names = source_names,
10 source_means = source_means,
11 source_sds = source_sds,
12 correction_means = correction_means,
13 correction_sds = correction_sds,
14 concentration_means = concentration_means
15 )
16)
17# Plot
18plot(simmr_1)
19# MCMC run
20simmr_1_out <- simmr_mcmc(simmr_1)
21# Plot
22plot(simmr_1_out) # Creates all 4 plots
23plot(simmr_1_out, type = "boxplot")
24plot(simmr_1_out, type = "histogram")
25plot(simmr_1_out, type = "density")
26plot(simmr_1_out, type = "matrix")
27## End(Not run)

描述
该函数获取simmr_mcmc的输出,并绘制后验预测分布,以实现模型拟合的可视化。模拟的后验预测值作为对象的一部分返回,并可以保存以供外部使用。
用法
1posterior_predictive(simmr_out, group = 1, prob = 0.5, plot_ppc = TRUE)
输入参数

示例
1data(geese_data_day1)
2simmr_1 <- with(
3 geese_data_day1,
4 simmr_load(
5 mixtures = mixtures,
6 source_names = source_names,
7 source_means = source_means,
8 source_sds = source_sds,
9 correction_means = correction_means,
10 correction_sds = correction_sds,
11 concentration_means = concentration_means
12 )
13)
14# Plot
15plot(simmr_1)
16# Print
17simmr_1
18# MCMC run
19simmr_1_out <- simmr_mcmc(simmr_1)
20# Prior predictive
21post_pred <- posterior_predictive(simmr_1_out)
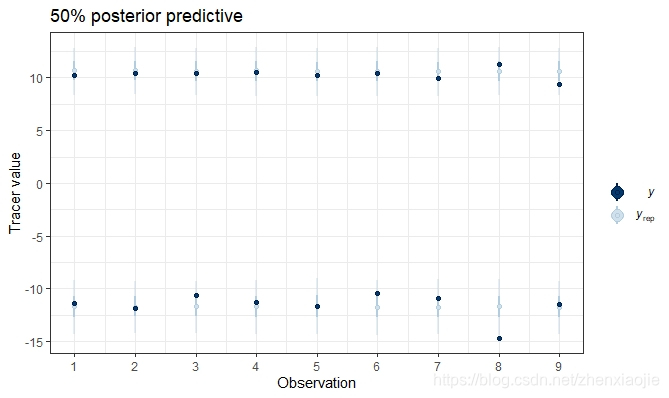

描述
打印simmr_input对象
用法
1## S3 method for class 'simmr_input'
2print(x, ...)
输入参数

输出值
一个简洁的simmr对象概要

描述
打印simmr_output对象
用法
1## S3 method for class 'simmr_output'
2print(x, ...)
输入参数

输出值
一个简洁的simmr对象概要

描述
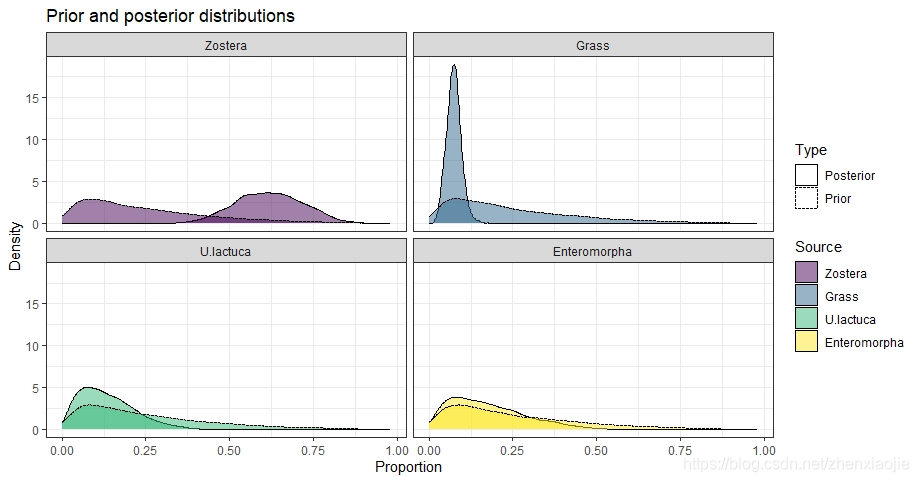
该函数获取simmr_mcmc的输出,并绘制先验分布,以支持目视检查。它可以单独使用,也可以作为posterior_predictive的一部分,直观地评估先验对后验分布的影响。
用法
1 prior_viz(
2 simmr_out,
3 group = 1,
4 plot = TRUE,
5 include_posterior = TRUE,
6 n_sims = 10000,
7 ggargs = NULL
8)
输入参数

示例
1## Not run:
2data(geese_data_day1)
3simmr_1 <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15# Plot
16plot(simmr_1)
17# Print
18simmr_1
19# MCMC run
20simmr_1_out <- simmr_mcmc(simmr_1)
21# Prior predictive
22prior <- prior_viz(simmr_1_out)
23head(prior)
24summary(prior)
25## End(Not run)
绘制先验及后验分布,以支持目视检查
先验输出的头几行信息
1> head(prior)
2 Zostera Grass U.lactuca Enteromorpha
3[1,] 0.10507054 0.64885067 0.15733823 0.08874057
4[2,] 0.50650097 0.08686476 0.37186878 0.03476549
5[3,] 0.01744883 0.01912078 0.87865924 0.08477114
6[4,] 0.14082734 0.73700084 0.07352376 0.04864805
7[5,] 0.45185621 0.13327463 0.19547573 0.21939342
8[6,] 0.26745860 0.02423665 0.26462609 0.44367865
1> summary(prior)
2 Zostera Grass U.lactuca
3 Min. :0.003194 Min. :0.002479 Min. :0.00195
4 1st Qu.:0.100435 1st Qu.:0.101082 1st Qu.:0.10143
5 Median :0.201689 Median :0.198843 Median :0.20160
6 Mean :0.249495 Mean :0.249668 Mean :0.25038
7 3rd Qu.:0.355536 3rd Qu.:0.355249 3rd Qu.:0.35498
8 Max. :0.977925 Max. :0.956716 Max. :0.96505
9 Enteromorpha
10 Min. :0.002751
11 1st Qu.:0.100769
12 Median :0.201069
13 Mean :0.250460
14 3rd Qu.:0.359518
15 Max. :0.958228

描述
这个包运行一个简单的稳定同位素混合模型(SIMM),可以作为对SIAR模型的一个长期替代。通过观察从生物体组织样本中获取的稳定同位素值,可以推断生物体的饮食比例。然而,SIMMs也可用于其他情况,例如在沉积物混合或脂肪酸组成中。主要函数是simmr_load和simmr_mcmc。帮助文件包含使用这个包的示例。更多信息请通过vignette()查询。
详细
正确运行SIMMs的一个更长期的替代是MixSIAR,它允许更详细的随机效应且包含协变量。
示例
1# A first example with 2 tracers (isotopes), 10 observations, and 4 food sources
2data(geese_data_day1)
3simmr_in <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15# Plot
16plot(simmr_in)
17# MCMC run
18simmr_out <- simmr_mcmc(simmr_in)
19# Check convergence - values should all be close to 1
20summary(simmr_out, type = "diagnostics")
21# Look at output
22summary(simmr_out, type = "statistics")
23# Look at influence of priors
24prior_viz(simmr_out)
25# Plot output
26plot(simmr_out, type = "histogram")
27## End(Not run)

描述
一个简单的假数据集,包括对2种同位素、4个来源、校正/营养富集因子(TEFs或TDFs)和浓度依赖均值的10个观测结果
用法
simmr_data_1
格式


描述
假数据集包含对3种同位素、4种来源、校正/营养富集因子(TEFs或TDFs)和浓度依赖均值的30次观测
用法
simmr_data_2
格式


描述
simmr_mcmc主要功能允许预先为膳食比例设置分布。先验分布是通过使用集中化对数比(CLR)来改变膳食比例来确定的。simmr_elicit和simmr_elicit函数允许用户指定每种膳食比例的先验平均值和标准差,然后找到适合输入simmr_mcmc的经CLR-转换的值。
用法
1simmr_elicit(
2 n_sources,
3 proportion_means = rep(1/n_sources, n_sources),
4 proportion_sds = rep(0.1, n_sources),
5 n_sims = 1000
6)
输入参数

详细
该函数获取所需的比例均值和标准偏差,并依次对均值和标准偏差拟合优化的最小二乘,从而生成用于simmr_mcmc的经CLR-转换的估计。考虑到推理的局限性,在SIMMs中使用先验信息是非常可取的。先验信息可能来自先前的研究、其他实验或其他对动物行为的观察。由于膳食比例所能跨越的空间有限,以及这个函数使用数值优化的事实,这个过程将不能完全匹配目标膳食比例的平均值和标准偏差。如果这个问题很严重,请尝试增加n_sims值。
输出值
两个列表对象

示例
1# Data set: 10 observations, 2 tracers, 4 sources
2data(geese_data_day1)
3simmr_1 <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15# MCMC run
16simmr_1_out <- simmr_mcmc(simmr_1)
17# Look at the prior influence
18prior_viz(simmr_1_out)
19# Summary
20summary(simmr_1_out, "quantiles")
geese_data_day1数据集分析的结果概要,以分位数(quantiles)形式展示1Summary for 1
2 2.5% 25% 50% 75% 97.5%
3deviance 51.252 52.668 54.141 56.254 62.608
4Zostera 0.421 0.549 0.617 0.687 0.817
5Grass 0.035 0.060 0.073 0.088 0.116
6U.lactuca 0.019 0.072 0.124 0.189 0.322
7Enteromorpha 0.022 0.082 0.151 0.239 0.427
8sd[d13C_Pl] 0.059 0.487 0.865 1.282 2.414
9sd[d15N_Pl] 0.014 0.162 0.366 0.632 1.457
1prior <- simmr_elicit(4, c(0.5, 0.2, 0.2, 0.1), c(0.08, 0.02, 0.01, 0.02))
2simmr_1a_out <- simmr_mcmc(simmr_1, prior_control = list(means = prior$mean, sd = prior$sd))
3#' # Look at the prior influence now
4prior_viz(simmr_1a_out)
5summary(simmr_1a_out, "quantiles")
1Summary for 1
2 2.5% 25% 50% 75% 97.5%
3deviance 56.046 59.860 62.323 64.817 70.388
4Zostera 0.541 0.599 0.628 0.657 0.705
5Grass 0.119 0.139 0.152 0.165 0.191
6U.lactuca 0.120 0.139 0.150 0.162 0.186
7Enteromorpha 0.042 0.057 0.067 0.079 0.109
8sd[d13C_Pl] 0.716 1.571 2.093 2.728 4.489
9sd[d15N_Pl] 0.015 0.152 0.341 0.583 1.243

描述
该函数接受混合物数据、食物源均值和标准偏差,(可选)校正因子均值和标准偏差,以及浓度比例。它对数据执行一些(非详尽的)检查,以确保它将通过simmr运行。它输出simmr_input类的对象。
用法
1simmr_load(
2 mixtures,
3 source_names,
4 source_means,
5 source_sds,
6 correction_means = NULL,
7 correction_sds = NULL,
8 concentration_means = NULL,
9 group = NULL
10)
输入参数

详细
对于标准稳定同位素混合物模型,混合物矩阵将包含的每行代表一个样本,每列代表一个同位素值。simmr可对在计算范围内的任何数量的同位素和任何观测次数进行分析。每种食物源的每种同位素都应提供源平均值/标准差。如果需要,校正方法(通常是营养富集因子)可以设为零,并且应与源值具有相同的形状。浓度依赖均值应为所述食物源中每种元素所占比例的估计数,并以0到1之间的比例形式给出。目前还没有包括浓度依赖标准差的方法。
输出值
simmr_input类的对象,包含以下元素:

示例
1# A simple example with 10 observations, 2 tracers and 4 sources
2data(geese_data_day1)
3simmr_1 <- with(
4 geese_data_day1,
5 simmr_load(
6 mixtures = mixtures,
7 source_names = source_names,
8 source_means = source_means,
9 source_sds = source_sds,
10 correction_means = correction_means,
11 correction_sds = correction_sds,
12 concentration_means = concentration_means
13 )
14)
15print(simmr_1)

描述
这是simmr包的主函数。它接受一个通过simmr_load创建的simmr_input对象,运行MCMC来确定饮食比例,然后输出一个simmr_output对象,以便通过summary.simmr_output和plot.simmr_output进行进一步分析和绘图。
用法
1simmr_mcmc(
2 simmr_in,
3 prior_control = list(means = rep(0, simmr_in$n_sources), sd = rep(1,simmr_in$n_sources)),
4 mcmc_control = list(iter = 10000, burn = 1000, thin = 10, n.chain = 4)
5)
输入参数

详细
如果,执行simmr_mcmc后,summary.simmr_output中的收敛诊断不能令人满意,mcmc_control中的iter、burn和thin的值应该增加10倍。
输出值
simmr_output类的一个对象,包含两个结构体命名的对象:

示例
1## See the package vignette for a detailed run through of these 4 examples
2# Data set 1: 10 obs on 2 isos, 4 sources, with tefs and concdep
3data(geese_data_day1)
4simmr_1 <- with(
5 geese_data_day1,
6 simmr_load(
7 mixtures = mixtures,
8 source_names = source_names,
9 source_means = source_means,
10 source_sds = source_sds,
11 correction_means = correction_means,
12 correction_sds = correction_sds,
13 concentration_means = concentration_means
14 )
15)
16# Plot
17plot(simmr_1)
18# Print
19simmr_1
20# MCMC run
21simmr_1_out <- simmr_mcmc(simmr_1)
22# Print it
23print(simmr_1_out)
24# Summary
25summary(simmr_1_out, type = "diagnostics")
26summary(simmr_1_out, type = "correlations")
27summary(simmr_1_out, type = "statistics")
28ans <- summary(simmr_1_out, type = c("quantiles", "statistics"))
29# Plot
30plot(simmr_1_out, type = "boxplot")
31plot(simmr_1_out, type = "histogram")
32plot(simmr_1_out, type = "density")
33plot(simmr_1_out, type = "matrix")
34# Compare two sources
35compare_sources(simmr_1_out, source_names = c("Zostera", "Enteromorpha"))
36# Compare multiple sources
37compare_sources(simmr_1_out)
1#####################################################################################
2# A version with just one observation
3data(geese_data_day1)
4simmr_2 <- with(
5 geese_data_day1,
6 simmr_load(
7 mixtures = mixtures[1, , drop = FALSE],
8 source_names = source_names,
9 source_means = source_means,
10 source_sds = source_sds,
11 correction_means = correction_means,
12 correction_sds = correction_sds,
13 concentration_means = concentration_means
14 )
15)
16# Plot
17plot(simmr_2)
18# MCMC run - automatically detects the single observation
19simmr_2_out <- simmr_mcmc(simmr_2)
20# Print it
21print(simmr_2_out)
22# Summary
23summary(simmr_2_out)
24summary(simmr_2_out, type = "diagnostics")
25ans <- summary(simmr_2_out, type = c("quantiles"))
26# Plot
27plot(simmr_2_out)
28plot(simmr_2_out, type = "boxplot")
29plot(simmr_2_out, type = "histogram")
30plot(simmr_2_out, type = "density")
31plot(simmr_2_out, type = "matrix")
1#####################################################################################
2# Data set 4 - identified by Fry (2014) as a failing of SIMMs
3# See the vignette for more interpreation of these data and the output
4# The data
5data(square_data)
6simmr_4 <- with(
7 square_data,
8 simmr_load(
9 mixtures = mixtures,
10 source_names = source_names,
11 source_means = source_means,
12 source_sds = source_sds
13 )
14)
15# Get summary
16print(simmr_4)
17# Plot
18plot(simmr_4)
19# MCMC run - needs slightly longer
20simmr_4_out <- simmr_mcmc(simmr_4)
21# Print it
22print(simmr_4_out)
23# Summary
24summary(simmr_4_out)
25summary(simmr_4_out, type = "diagnostics")
26ans <- summary(simmr_4_out, type = c("quantiles", "statistics"))
27# Plot
28plot(simmr_4_out)
29plot(simmr_4_out, type = "boxplot")
30plot(simmr_4_out, type = "histogram")
31plot(simmr_4_out, type = "density")
32plot(simmr_4_out, type = "matrix") # Look at the massive correlations here
1#####################################################################################
2# Data set 4 - identified by Fry (2014) as a failing of SIMMs
3# See the vignette for more interpreation of these data and the output
4# The data
5data(square_data)
6simmr_4 <- with(
7 square_data,
8 simmr_load(
9 mixtures = mixtures,
10 source_names = source_names,
11 source_means = source_means,
12 source_sds = source_sds
13 )
14)
15# Get summary
16print(simmr_4)
17# Plot
18plot(simmr_4)
19# MCMC run - needs slightly longer
20simmr_4_out <- simmr_mcmc(simmr_4)
21# Print it
22print(simmr_4_out)
23# Summary
24summary(simmr_4_out)
25summary(simmr_4_out, type = "diagnostics")
26ans <- summary(simmr_4_out, type = c("quantiles", "statistics"))
27# Plot
28plot(simmr_4_out)
29plot(simmr_4_out, type = "boxplot")
30plot(simmr_4_out, type = "histogram")
31plot(simmr_4_out, type = "density")
32plot(simmr_4_out, type = "matrix") # Look at the massive correlations here
1#####################################################################################
2# Data set 5 - Multiple groups Geese data from Inger et al 2006
3# Do this in raw data format - Note that there's quite a few mixtures!
4data(geese_data)
5simmr_5 <- with(
6 geese_data,
7 simmr_load(
8 mixtures = mixtures,
9 source_names = source_names,
10 source_means = source_means,
11 source_sds = source_sds,
12 correction_means = correction_means,
13 correction_sds = correction_sds,
14 concentration_means = concentration_means,
15 group = groups
16 )
17)
18# Plot
19plot(simmr_5,
20 xlab = expression(paste(delta^13, "C (\\u2030)", sep = "")),
21 ylab = expression(paste(delta^15, "N (\\u2030)", sep = "")),
22 title = "Isospace plot of Inger et al Geese data"
23)
24# Run MCMC for each group
25simmr_5_out <- simmr_mcmc(simmr_5)
26# Summarise output
27summary(simmr_5_out, type = "quantiles", group = 1)
28summary(simmr_5_out, type = "quantiles", group = c(1, 3))
29summary(simmr_5_out, type = c("quantiles", "statistics"), group = c(1, 3))
30# Plot - only a single group allowed
31plot(simmr_5_out, type = "boxplot", group = 2, title = "simmr output group 2")
32plot(simmr_5_out, type = c("density", "matrix"), grp = 6, title = "simmr output group 6")
33# Compare sources within a group
34compare_sources(simmr_5_out, source_names = c("Zostera", "U.lactuca"), group = 2)
35compare_sources(simmr_5_out, group = 2)
36# Compare between groups
37compare_groups(simmr_5_out, source = "Zostera", groups = 1:2)
38compare_groups(simmr_5_out, source = "Zostera", groups = 1:3)
39compare_groups(simmr_5_out, source = "U.lactuca", groups = c(4:5, 7, 2))
40## End(Not run)

描述
该函数运行一个与simmr_mcmc主函数略有不同的版本,主要区别在于,对于一个给定的膳食比例,它估计了校正因子(有时称为营养富集或营养区分因子;TEFs/TDFs)。
用法
1simmr_mcmc_tdf(
2 simmr_in,
3 p = matrix(rep(1/simmr_in$n_sources, simmr_in$n_sources), ncol = simmr_in$n_sources,
4 nrow = simmr_in$n_obs, byrow = TRUE),
5 prior_control = list(c_mean_est = rep(2, simmr_in$n_tracers), c_sd_est = rep(2,
6 simmr_in$n_tracers)),
7 mcmc_control = list(iter = 10000, burn = 1000, thin = 10, n.chain = 4)
8)
输入参数

详细
simmr_load创建的对象,其中包含一些示踪剂和食物源均值和标准偏差的混合数据。该对象中包含的任何修正因素将被忽略。应该为每个个体提供已知的膳食比例(即应该是一个与mix相同的行数的矩阵)。建议在饲喂试验设计中采用多种不同膳食比例。simmr_mcmc的输出一样,应该检查它是否收敛。下面包含了一些示例,以帮助进行检查和进一步绘图。simmr_mcmc_tdf后,summary.simmr_output_tdf中的收敛诊断不令人满意,mcmc_control中iter、burn和thin的值应该增加10倍。输出值
simmr_output类的一个对象,包含两个结构体命名的对象:

示例
1# Data set 1: 10 obs on 2 isos, 4 sources, with tefs and concdep
2# Assume p = c(0.25, 0.25, 0.25, 0.25)
3# The data
4data(simmr_data_1)
5# Load into simmr
6simmr_tdf <- with(
7 simmr_data_1,
8 simmr_load(
9 mixtures = mixtures,
10 source_names = source_names,
11 source_means = source_means,
12 source_sds = source_sds,
13 correction_means = correction_means,
14 correction_sds = correction_sds,
15 concentration_means = concentration_means
16 )
17)
18# Plot
19plot(simmr_tdf)
20# MCMC run
21simmr_tdf_out <- simmr_mcmc_tdf(simmr_tdf,
22 p = matrix(rep(
23 1 / simmr_tdf$n_sources,
24 simmr_tdf$n_sources
25 ),
26 ncol = simmr_tdf$n_sources,
27 nrow = simmr_tdf$n_obs,
28 byrow = TRUE
29 )
30)
31# Summary
32summary(simmr_tdf_out, type = "diagnostics")
33summary(simmr_tdf_out, type = "quantiles")
34# Now put these corrections back into the model and check the
35# iso-space plots and dietary output
36simmr_tdf_2 <- with(
37 simmr_data_1,
38 simmr_load(
39 mixtures = mixtures,
40 source_names = source_names,
41 source_means = source_means,
42 source_sds = source_sds,
43 correction_means = simmr_tdf_out$c_mean_est,
44 correction_sds = simmr_tdf_out$c_sd_est,
45 concentration_means = concentration_means
46 )
47)
48# Plot with corrections now
49plot(simmr_tdf_2)
50simmr_tdf_2_out <- simmr_mcmc(simmr_tdf_2)
51summary(simmr_tdf_2_out, type = "diagnostics")
52plot(simmr_tdf_2_out, type = "boxplot")

描述
作为SIMMs分析的失败案例,由Fry(2014)定义的假箱式数据集,请查看链接了解更多的对该数据和输出的解读。
用法
square_data
数据集格式
一个包含下列元素的列表

来源
https://www.int-res.com/abstracts/meps/v490/p285-289/

描述
为使用simmr_mcmc创建的对象生成文本摘要和收敛诊断。不同的选项是:“诊断”产生Brooks-Gelman-Rubin诊断来评估MCMC收敛性,“分位数”产生参数的置信区间,“统计”产生平均值和标准偏差,以及“相关性”产生参数之间的相关性。
用法
1## S3 method for class 'simmr_output'
2summary(
3 object,
4 type = c("diagnostics", "quantiles", "statistics", "correlations"),
5 group = 1,
6 ...
7)
输入参数

详细
分位数输出可以方便地计算出95%可信区间的后验膳食比例。相关性、以及plot.simmr_output中的矩阵图,允许用户判断哪些源是不可识别的。Gelman诊断值应接近1,以保证满意的收敛。
输出值
包含以下组成的列表:

注意,该对象是静默报告的,因此将被丢弃,除非使用如下例中的对象调用该函数。
示例
1# A simple example with 10 observations, 2 tracers and 4 sources
2# The data
3data(geese_data_day1)
4simmr_1 <- with(
5 geese_data_day1,
6 simmr_load(
7 mixtures = mixtures,
8 source_names = source_names,
9 source_means = source_means,
10 source_sds = source_sds,
11 correction_means = correction_means,
12 correction_sds = correction_sds,
13 concentration_means = concentration_means
14 )
15)
16# Plot
17plot(simmr_1)
18# MCMC run
19simmr_1_out <- simmr_mcmc(simmr_1)
20# Summarise
21summary(simmr_1_out) # This outputs all the summaries
22summary(simmr_1_out, type = "diagnostics") # Just the diagnostics
23# Store the output in an
24ans <- summary(simmr_1_out,
25 type = c("quantiles", "statistics")
26)

描述
为使用simmr_mcmc_tdf创建的对象生成文本摘要和收敛诊断。不同的选项是:“诊断”产生Brooks-Gelman-Rubin诊断来评估MCMC收敛性,“分位数”产生参数的可信区间,“统计”产生平均值和标准偏差,以及“相关性”产生参数之间的相关性。
用法
1## S3 method for class 'simmr_output_tdf'
2summary(
3 object,
4 type = c("diagnostics", "quantiles", "statistics", "correlations"),
5 ...
6)
输入参数

详细
分位数输出可以方便地计算出95%可信区间的后验膳食比例。Gelman诊断值应接近1,以保证满意的收敛。目前不支持多个组来估计TDFs。
输出值
包含以下组成的列表:

注意,该对象是静默报告的,因此将被丢弃,除非使用如下例中的对象调用该函数。

江苏天诺基业生态科技有限公司 http://www.tynoocorp.com
滔涛云 http://taotaoyun.net
由香港宝典宝典免费资料大全甄晓杰提供相关新闻稿